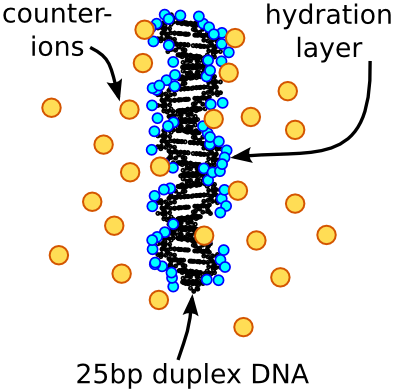DNA and RNA Electrostatics
Nucleic acids are highly charged, attracting a cloud of ions from the solutions that surround them. Changes in the ion atmosphere are important to consider in protein-DNA interactions and RNA folding. However, the structure and composition of the ion atmosphere is difficult to predict and measure.
X-ray solution scattering is uniquely sensitive to the diffuse ion clouds surrouding nucleic acids. While in the Pollack lab at Cornell, I used the anomalous scattering contrast of ionic species to count the ions around DNA and RNA and determine their spatial distribution (Pabit et al. NAR 2009 and Pabit et al. JACS 2010). I also worked with theorists to improve ion atmosphere modeling. With David Case and his group at Rutgers, we showed that accurate ion atmosphere predictions can be made with reasonable computational cost using implicit solvent models (Nguyen et al. J. Chem. Phys. 2014). In addition, I worked with Serdal Kirmizialtin and Ron Elber (UT Austin) to experimentally validate all-atom molecular dynamics simulations of RNA duplex and pseudoknot structures (Kirmizialtin et al. Biophys. J. 2012).
Highly charged polyions such as RNA and DNA can exist as expanded random coils; these coils resist collapse because like charges repel. If the charges are compensated by solutes (cations such as Na+, K+, or Mg2+), the molecules may form higher-order structures important for biological function: RNA folds and DNA hybridizes. Interestingly, the response of the random coil states to particular solutes has proven difficult to predict. Two techniques, single-molecule Förster Resonance Energy Transfer (smFRET) and solution X-ray scattering (SAXS), give complementary views of the conformational ensemble in solution, and have proven very powerful when applied in combination. As a member of the Pollack lab, I used these techniques to measure changes in the random coil state of RNA and DNA (Chen et al. PNAS 2012) as well as chemically-unfolded proteins (Yoo et al. J. Mol. Biol. 2012). In order to extract structural parameters from these measurements, we developed new computational tools, including a coarse-grained polyelectrolyte model for single-stranded DNA (Meisburger et al. Biopolymers 2013) and software for atomistic ensemble generation and fitting (Plumridge et al. NAR 2017).