Correlated motions in structural biology
Xu D†, Meisburger SP†, Ando N.
†equal contribution
Biochemistry 60: 2331–2340 (2021).
Abstract
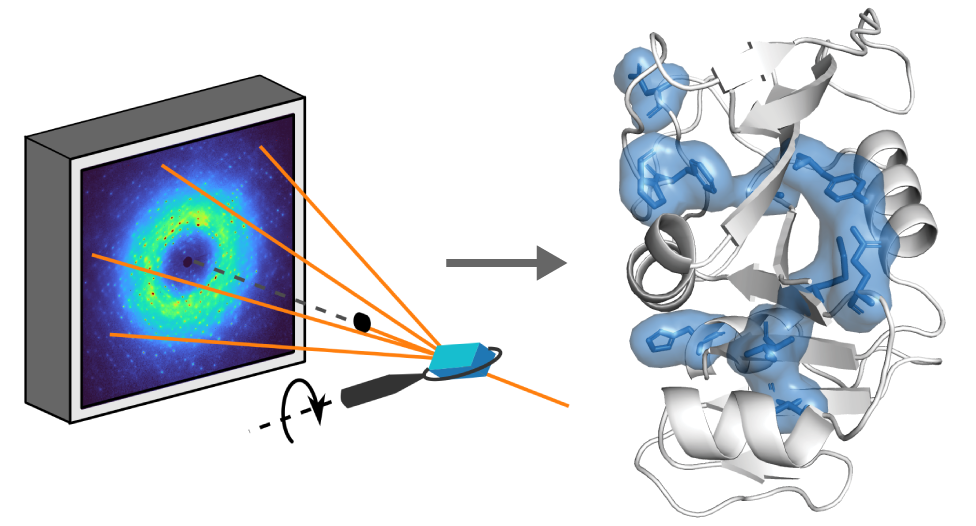
Correlated motions in proteins arising from the collective movements of residues have long been proposed to be fundamentally important to key properties of proteins, from allostery and catalysis to evolvability. Recent breakthroughs in structural biology have made it possible to capture proteins undergoing complex conformational changes, yet intrinsic correlated motions within a conformation remain one of the least understood facets of protein structure. For many decades, the analysis of total X-ray scattering held the promise of animating crystal structures with correlated motions. With recent advances in both X-ray detectors and data interpretation methods, this long-held promise can now be met. In this Perspective, we will introduce how correlated motions are captured in total scattering and provide guidelines for the collection, interpretation, and validation of data. As structural biology continues to push the boundaries, we see an opportunity to gain atomistic insight into correlated motions using total scattering as a bridge between theory and experiment.